Reprinted with permission of American Journal of Pathology, Bethesda, MD.
As published in American Journal
of Pathology Vol. 150, No. 6, pp. 2153-2165, 1997
Bradley J. Quade*,
Catherine M. McLachlin+, Malena Soto-Wright#, James
Zuckerman#, George L. Mutter*, and Cynthia C. Morton *#
From the Departments of Pathology* and Obstetrics, Gynecology, and
Reproductive Biology, # Brigham and Women's Hospital and Harvard
Medical School, Boston, Massachusetts, and the Department of Pathology, +
Victoria Hospital and University of Western Ontario, London, Ontario, Canada
Disseminated peritoneal
leiomyomatosis (DPL, leiomyomatosis peritonealis disseminata) is a rare
condition in which multiple histologically benign smooth muscle tumorlets
diffusely stud peritoneal and omental surfaces in females, predominantly of
reproductive age. Although the distribution of these lesions suggests a
metastatic process, DPL generally has a benign clinical course and has been
regarded as a metaplastic process. We assessed clonality of 42 tumorlets and 15
normal tissues from four females with DPL by analyzing X chromosome inactivation
as indicated by the methylation status of the androgen receptor gene (HUMARA).
In each of the four patients, the same parental X chromosome was non randomly
inactivated in all tumorlets, consistent with a metastatic unicentric neoplasm,
or alternatively, selection for an X-linked allele in clonal multicentric
lesions. Anomalous demethylation of the marker for X inactivation (HUMARA) was
associated with loss of heterozygosity for markers spanning the X chromosome, or
monosomy X, in part of one leiomyomatous lesion. Biallelic demethylation of the
HUMARA microsatellite polymorphism was also found in one intramural lyomyoma.
Two of six DPL lesions karyotyped had cytogenetic abnormalities involving
chromosomes 7, 12, and 18, suggesting a pathogenesis in common with uterine
leiomyomas. (Am J Pathol 1997, 150:2153-2166)
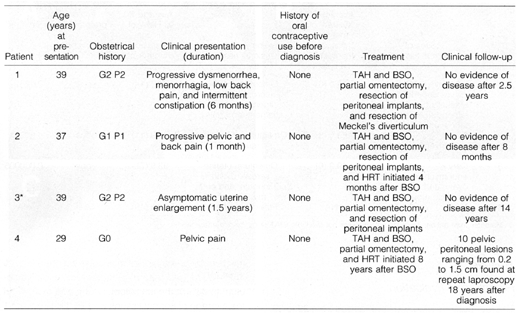
Table 1. Clinical Features of Four Patients
with DPL

TAH, total abdominal hystorectomy;
BSO, bilateral salpingo-oopherectomy; HRT, hormone replacement therapy.
*Previously reported.15
Table 2. Nonrandom X Chromosome Inactivation
in DPL
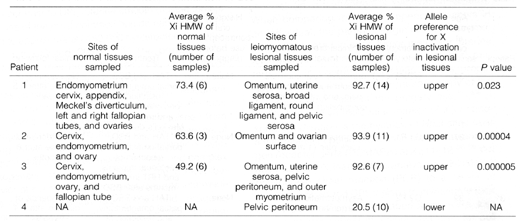
Materials from patient 4's
hysterectomy were not available (NA) for our review and analysis.
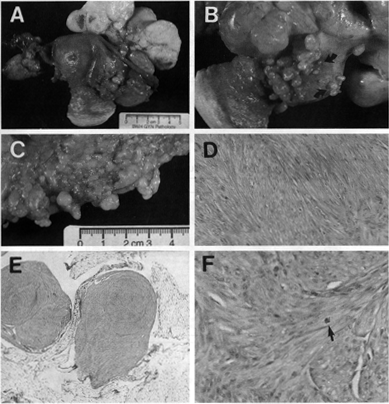
Figure 1. Gross and
microscopic pathological features of DPL. The cut surface of a large
leiomyomatous mass that arose in the round ligament of patient 1 was firm,
white, bulging, whorled, and focally hemorrhagic (A). Innumerable smaller
leiomyomatous lesions ranging from 0.01 to 2.0 cm in greatest dimension studded
the peritoneal surfaces of the anterior uterus, broad ligaments, para-ovarian
soft tissue (A), posterior uterus (B), and omentum (C and E).
These nodular lesions contained interwoven fascicles of smooth muscle cells with
mild to moderate hypercellulatity and without coagulative tumor necrosis or
cytological or nuclear atypia (D and E). Mitotic figures were rare
(<2 per 100 high-powerfields in 300 fields counted; arrow in F). Atypical
mitotic figures were not present. For comparison, uterine smooth muscle tumors
without significant atypia or necrosis are generally not considered to have an
uncertain malignant potential until the mitotic index is greater than 5 mitotic
figures per 10 high-powerfields.32,31,84 In all four patients, microscopic foci
of endometrial epithelium and stroma were present in association with
leiomyomatous proliferations and resembled endometriosis (not shown). In patient
2, a microscopic cyst lined by histologically benign simple endocervical-type
epithelium was found in one leiomyomatous nodule (not shown).
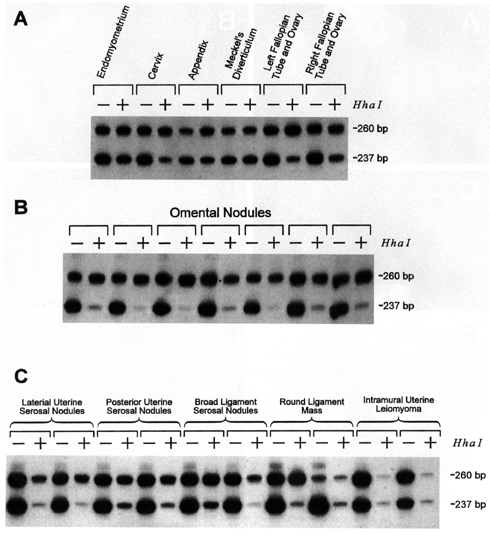
Figure 2.Analysis of
clonality by PCR amplification of the HUMARA microsatellite polymorpbism with
(+) and without (-) previous HhaI restriction digestion. DNA was extracted from
paraffin-embedded normal (A), omental (B), and non-omental
peritoneal (C) leiomyomatous nodules from patient 1. A: Mildly skewed X
chromosome inactivation was found in normal tissues. B and C:
Nonrandom X inactivation was found in all leiomyomatous lesions. C:
Anomalous patterns of HUMARA PCR were found in one of two samples from the large
round ligament mass and in both samples from the intramural uterine leiomyoma.
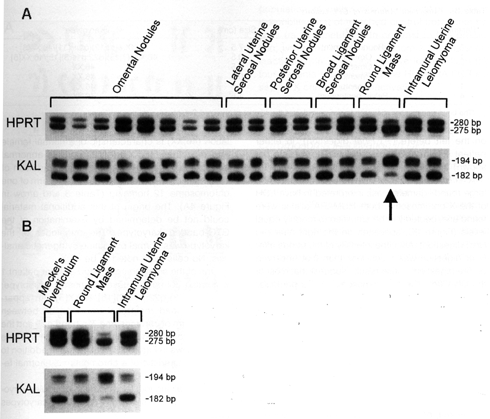
Figure 3. Evidence of focal
loss of heterozygosity (A) and DNA methylation (B) of the X
chromosome in leiomyomatous lesions in patient 1. Paraffin-extracted DNA from
patient 1 was amplified using primers to the HPRTB locus at Xq26.1 and the KAL
locus at Xp22.3. In A, LOH for the markers spanning the entire X
chromosome was found in one of two samples (arrow) from the 7.5-cm round
ligament mass shown in Figure 1A. LOH for the X chromosome was not found in
eight omental and seven other extra-omental leiomyomatous lesions. In B,
DNA from samples (one from the round ligament mass and one from the intramural
leiomyoma) with anomalous results for PCR amplification at the HUMARA locus were
amplified with primers for HPRTB and KAL after previous HhaI digestion. These
anomalous samples were compared with HhaI-digested DNA from the other round
ligament mass sample (lacking LOH at HUMARA) and one control tissue (Meckel's
diverticulum) with a polygonal pattern at the HUMARA locus. The lack of
amplification at HUMARA, but not HPRTB and KAL, after HhaI digestion is
consistent with an epigenetic loss of DNA methylation at HUMARA in the
intramural leiomyoma.
Table 3. Cytogenetic Analysis of DPL Lesions
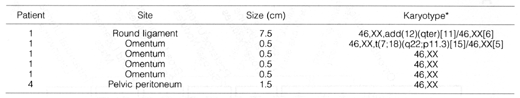
*The bracketed number after each
karyotype indicates the number of analyzed metaphases with that karyotype.
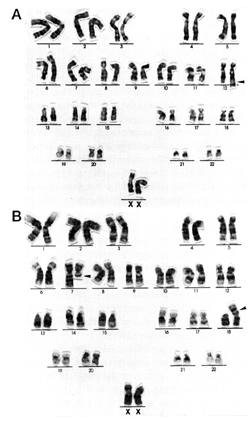
Figure 4. Cytogenetic abnormalities in DPL patient 1 included structural rearrangements of chromosomes 12 (A) and 7 and 18 (B). The karyotype of the large round ligament leiomyoma (Figure 1A) was 46,XX,add(12) (qter)[11]/46,XX[6]. The additional chromatin on cbromosome 12 is indicated by the arrowhead in A. The abnormal karyotype of an omental nodule was 46,XX,t(7;18)(q22;p11.3)[15]/46,XX[5]. The translocation is identified by the arrowheads in panel B. The bracketed number following each karyotype indicates the number of analyzed metaphases with that karyotype.